ORIGINAL ARTICLE
In silico analysis of potential loci for the identification of Vanda spp. in the Philippines
1
Department of Biology, College of Science, Bulacan State University,
Malolos, Bulacan, Philippines 3000
2
Department of Biology, College of Science, De La Salle University,
Manila, Philippines
Online publication date: 2023-07-31
Publication date: 2023-07-31
Plant and Fungal Systematics 2023; 68(1): 223-231
KEYWORDS
ABSTRACT
Difficulties in identifying Vanda species are still encountered, and the ambiguity
in its taxonomy is still unresolved. To date, the advancement in molecular genetics
technology has given rise to the molecular method for plant identification and elucidation.
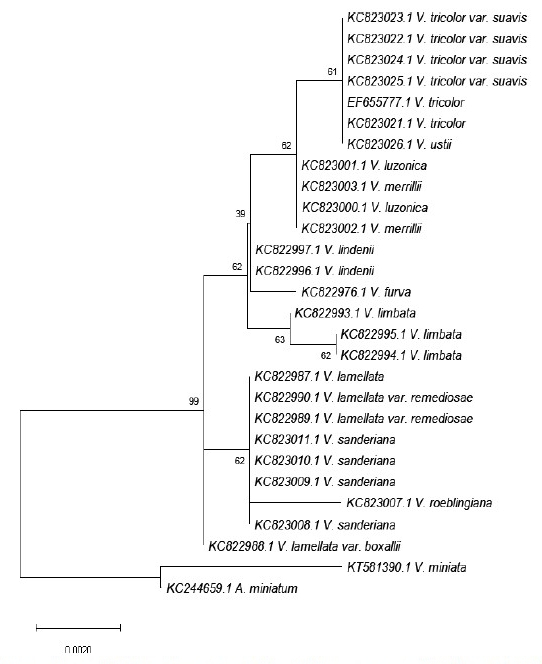
One hundred twenty-five (125) gene sequences of Vanda species from the Philippines
were obtained from the NCBI GenBank. Four of the 25 loci were further examined using
MEGA 11 software for multiple sequence alignment, sequence analysis, and phylogenetic
reconstruction. The indel-based and tree-based methods were combined to compute the
species resolution. The result showed that ITS from the nuclear region obtained the highest
species resolution with 66.67%. It was then followed by psbA-trnH, matK, and trnL-trnF
from the chloroplast genome with a species resolution of 60%, 40%, and 30.77%, respectively.
ITS and psbA-trnH satisfied the ideal length for DNA barcoding as they have 655 bp
and 701 bp, respectively. The locus psbA-trnH was also considered to have a higher potential
to discriminate Vanda species since only a few sequences were tested for ITS. Furthermore,
ITS and trnL-trnF have the highest variable rate, which is 2.9%, while matK and psbA-trnH
have 2% and 1.3%, respectively. This showed the nature of the unique sequences
of various species. In this study, the indel-based method provided better results than the
tree-based method. It will help support further DNA barcoding studies and strengthen the
conservation and protection of Vanda spp. in the Philippines.
FUNDING
NO
REFERENCES (21)
1.
Chatzou, M., Magis, C., Chang, J. M., Kemena, C., Bussotti, G., Erb, I. & Notredame, C. 2016. Multiple sequence alignment modeling: methods and applications. Briefings in Bioinformatics 17(6): 1009–1023. https://doi.org/10.1093/bib/bb....
2.
DENR Administrative Order 2017. Updated national list of threatened Philippine plants and their categories. (DAO 2017-11). https://bmb.gov.ph/index.php/f.... Access date: February 2022.
3.
Dizon, S. A., Ocenar, A. P. & Naïve, M. A. K. 2018. Inventory of orchids in the Mount Hamiguitan Range Wildlife Sanctuary, Davao Oriental, Philippines. Bio Bulletin 4(1): 37–42.
4.
Edgar, R. C. 2004a. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5): 1792–1797. https://doi.org/10.1093/nar/gk....
5.
Edgar, R. C. 2004b. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics 5: 113.
6.
Gardiner, L. M., Kocyan, A., Motes, M., Roberts, D. L. & Emerson, B. C. 2013. Molecular phylogenetics of Vanda and related genera (Orchidaceae). Botanical Journal of the Linnean Society 173(4): 549–572. https://doi.org/10.1111/BOJ.12....
7.
Hall, B. G. 2013. Building phylogenetic trees from molecular data with MEGA. Molecular Biology and Evolution 30(5): 1229–1235. https://doi.org/10.1093/molbev....
8.
Hollingsworth, P. M., Graham, S. W. & Little, D. P. 2011. Choosing and using a plant DNA barcode. PLOS One 6(5): e19254. https://doi.org/10.1371/journa....
9.
Kim, H. M., Oh, S. H., Bhandari, G. S., Kim, C. S. & Park, C. W. 2013. DNA barcoding of Orchidaceae in Korea. Molecular Ecology Resources 14(3): 499–507. https://doi.org/10.1111/1755-0....
10.
Kinene, T., Wainaina, J., Maina, S. & Boykin, L. M. 2016. Rooting trees, methods for. Encyclopedia of Evolutionary Biology 13: 489–493. https://doi.org/10.1016/B978-0....
11.
Lahaye, R., Van der Bank, M., Bogarin, D., Warner, J., Pupulin, F., Gigot, G. & Savolainen, V. 2008. DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences 105(8): 2923–2928. https://doi.org/10.1073/pnas.0....
12.
Nguyen, N. H., Vu, H. T., Le, N. D., Nguyen, T. D., Duong, H. X. & Tran, H. D. 2020. Molecular identification and evaluation of the genetic diversity of dendrobium species collected in southern Vietnam. Biology 9(4): 76. https://doi.org/10.3390/biolog....
13.
Panal, C. L. T., Opiso, J. G. & Opiso, G. 2015. Conservation status of the Family Orchidaceae in Mt. Sinaka, Arakan, North Cotabato, Philippines. Biodiversitas Journal of Biological Diversity 16: 213–224. https://doi.org/10.13057/biodi....
14.
Sanyal, G., Mahadani, A. K., Mahadani, P. & Bhattacharjee, P. 2015. Insertion-deletion as informative characters in DNA barcoding. International Journal of Multimedia and Ubiquitous Engineering 10(10): 67–74. https://doi.org/10.14257/ijmue....
15.
Tamura, K., Dudley, J., Nei, M. & Kumar, S. (n.d.). Molecular Evolutionary Genetics Analysis version 4. http://cda.psych.uiuc.edu/406_.... Access date: January 2022.
16.
Tamura, K., Stecher, G. & Kumar, S. 2021. MEGA 11: Molecular Evolutionary Genetics Analysis version 11. Molecular Biology and Evolution 38(7): 3022–3027. https://doi.org/10.1093/molbev....
17.
Tan, G., Muffato, M., Ledergerber, C., Herrero, J., Goldman, N., Gil, M. & Dessimoz, C. 2015. Current methods for automated filtering of multiple sequence alignments frequently worsen single-gene phylogenetic inference. Systematic Biology 64(5): 778–791. https://doi.org/10.1093/sysbio....
18.
Tanee, T., Chadmuk, P., Sudmoon, R., Chaveerach, A. & Noikotr, K. 2012. Genetic analysis for identification, genomic template stability in hybrids and barcodes of the Vanda species (Orchidaceae) of Thailand. African Journal of Biotechnology 11(55): 11772–11781. https://doi.org/10.5897/AJB11.....
19.
Vu, T. H. T, Le, T. L., Nguyen, T. K., Tran, D. D. & Tran, H. D. 2017. Review on molecular markers for identification of Orchids. Vietnam Journal of Science, Technology and Engineering 59(2): 62–75. https://doi.org/10.31276/VJSTE....
20.
Vu, H. T., Huynh, P., Tran, H. D. & Le, L. 2018. In silico study on molecular sequences for identification of Paphiopedilum species. Evolutionary Bioinformatics. https://doi.org/10.1177/117693....
21.
Vu, H. T., Vu, Q. L., Nguyen, T. D., Tran, N., Nguyen, T. C., Luu, P. N. & Le, L. 2020. Genetic diversity and identification of Vietnamese Paphiopedilum species using DNA Sequences. Biology 9(1): 9. https://doi.org/10.3390/biolog....
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.