A new species of Tetradesmus (Chlorophyceae, Chlorophyta)
isolated from desert soil crust habitats in southwestern
North America
1
Department of Ecology and Evolutionary Biology, University of Connecticut,
75 North Eagleville Rd., Storrs, CT 06269-3043, USA
Publication date: 2019-07-18
Plant and Fungal Systematics 2019; 64(1): 25-32
KEYWORDS
ABSTRACT
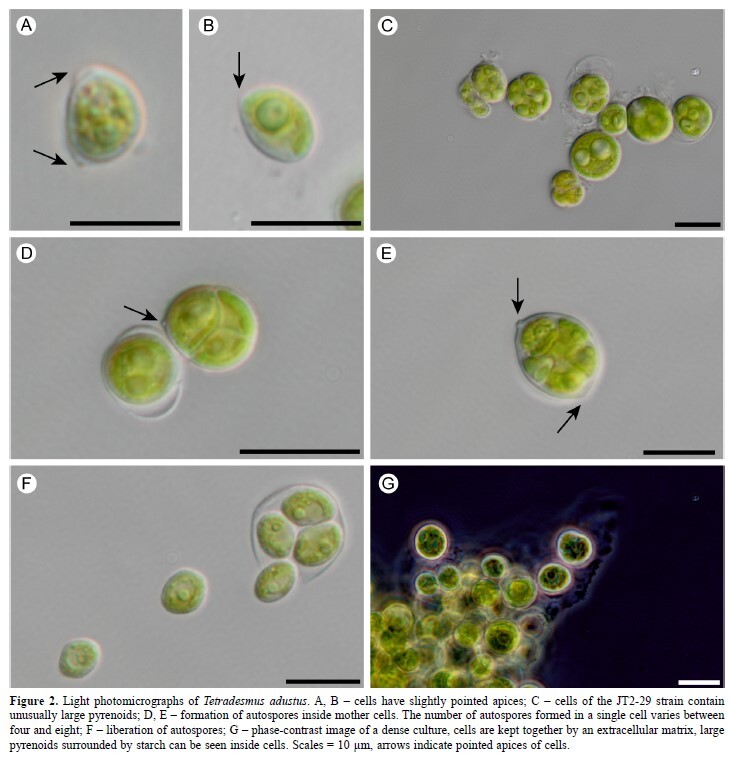
A new species of Tetradesmus (Tetradesmus adustus) is described from desert
soils of southwestern North America. The identification is based on phylogenetic analysis
of data from nuclear (ITS2 rDNA) and plastid (rbcL, tufA) barcode markers. This newly
described species represents the fifth cryptic species of arid-adapted algae in Scenedesmaceae.
A re-analysis of published sequences attributed to desert Tetradesmus in the context
of our newly obtained data reiterates the importance of robust phylogenetic analysis in
identification of cryptic taxa, such as species of Tetradesmus.
REFERENCES (41)
1.
An, S. S., Friedl, T. & Hegewald, E. 1999. Phylogenetic relationships of Scenedesmus and Scenedesmus-like coccoid green algae as inferred from ITS-2 rDNA sequence comparisons. Plant Biology 1: 418–428.
2.
Belnap, J. & Lange, O. L. 2001. Biological soil crusts: structure, function, and management. Springer, Berlin.
3.
Bold, H. C. 1949. The morphology of Chlamydomonas chlamydogama, Sp. Nov. Bulletin Torrey botanical club 76: 101–108.
4.
Büdel, B., Darienko, T., Deutschewitz, K., Dojani, S., Friedl, T., Mohr, K. I., Salisch, M., Reisser, W. & Weber, B. 2009. Southern African biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Soil Microbiology 57: 229–247.
5.
Cameron, R. E. 1960. Communities of soil algae occurring in the Sonoran desert in Arizona. Journal of Arizona Academy of Science 1: 85–88.
6.
Cameron, R. E. 1964. Algae of Southern Arizona. II. Algal Flora (Exclusive of Bluegreen Algae). Review Algology 7: 151–177.
7.
Cardon, Z. G., Peredo, E. L., Dohnalkova, A. C., Gershone, H. L. & Bezanilla, M. 2018. A model suite of green algae within the Scenedesmaceae for investigating contrasting desiccation tolerance and morphology. Journal of Cell Science 131: jcs212233.
8.
De Clerck, O., Guiry, M. D., Leliaert, F., Samyn, Y. & Verbruggen, H. 2013. Algal taxonomy: a road to nowhere? Journal of Phycology 49: 215–225.
9.
Evans, R. D. & Johansen, J. R. 1999. Microbiotic crusts and ecosystem pocesses. Critical Reviews in Plant Sciences 18: 183–225.
10.
Famá, P., Wysor, B., Kooistra, W. & Zuccarello, G. C. 2012. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. Journal of Phycology 38: 1040–1050.
11.
Flechtner, V. R., Johansen, J. R. & Clark, W.H. 1998. Algal composition of microbiotic crusts from the Central Desert in Baja California, Mexico. Great Basin Naturalist 58: 295–311.
12.
Fučíková, K., Lewis, P. O. & Lewis, L. A. 2014. Widespread desert affiliation of trebouxiophycean Algae (Trebouxiophyceae, Chlorophyta) including discovery of three new desert genera. Phycological Research 62: 294–305.
13.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321.
14.
Hall, J., Fučíková, K., Lo, C., Lewis, L. A. & Karol, K. 2010. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogamie Algologie 31: 529–555.
15.
Huss, V. A. R., Frank, C., Hartmann, E. C., Hirmer. M., Kloboucek, A., Seidel, B. M., Wenzeler, P. & Kessler, E. 1999. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta). Journal of Phycology 35: 587–598.
16.
Koetschan, C., Hackl, T., Müller, T., Wolf, M., Förster, F. & Schultz, J. 2012. ITS2 database IV: interactive taxon sampling for Internal Transcribed Spacer 2 based phylogenies. Molecular Phylogenetics and Evolution 63: 585–588.
17.
Lanfear, R., Calcott, B., Ho, S. Y. & Guindon, S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701.
18.
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773.
19.
Lange, O. L., Kidron, G. J., Büdel, B., Meyer, A., Kilian, E. & Abeliovich, A. 1992. Taxonomic composition and photosynthetic characteristics of the biological soil crusts covering sand dunes in the Western Negev Desert. Functional Ecology 56: 519–527.
20.
Lewis, L. A. & Flechtner, V. R. 2004. Cryptic species of Scenedesmus (Chlorophyta) from desert soil communities of Western North America. Journal of Phycology 40: 1127–1137.
21.
Lewis, L. A. & Flechtner, V. R. 2019. Tetradesmus bajacalifornicus L.A.Lewis & Flechtner, sp. nov. and Tetradesmus deserticola L.A.Lewis & Flechtner, sp. nov. (Scenedesmaceae, Chlorophyta). Notulae algarum No. 88.
22.
Lewis, L. A. & Lewis, P. O. 2005. Unearthing the molecular phylodiversity of desert soil green algae (Chlorophyta). Systematic Biology 54: 936–947.
23.
Lürling, M. & Van Donk, E. 1996. Zooplankton-induced unicell-colony transformation in Scenedesmus acutus and its effect on growth of herbivore Daphnia. Oecologia 108: 432–437.
24.
Mikhailyuk, T., Glaser, K., Tsarenko, P., Demchenko, E. & Karsten, U. 2018. Composition of biological soil crusts from sand dunes of the Baltic Sea coast, in the context of an integrative approach to the taxonomy of microalgae and cyanobacteria. European Journal of Phycology. In press.
25.
Miller, M. A., Pfeiffer, W. & Schwartz, T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop 14 Nov. 2010, New Orleans, LA, pp. 1–8.
26.
Muggia, L., Leavitt, S. & Barreno, E. 2018. The hidden diversity of lichenised Trebouxiophyceae (Chlorophyta). Phycologia 57: 503–524.
27.
Ocampo-Paus, R. & Friedmann, I. 1966. Radiosphaera Negevensis Sp. N., a new chlorococcalean desert alga. American Journal of Botany 53: 663–671.
28.
Rambout, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904.
29.
Ronquist, F. & Huelsenbeck, J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
30.
Schultz, J., Müller, T., Achtziger, M., Seibel, P. N., Dandekar, T. & Wolf, M. 2006. The Internal Transcribed Spacer 2 Database – a web server for (not only) low level phylogenetic analyses. Nucleic Acids Research 34: 704–707.
31.
Schwarz, G. 1978. Estimating the dimension of a model. The Annals of Statistics 6: 461–464.
32.
Sciuto, K., Lewis, L. A., Verleyen, E., Moro, I. & La Rocca, N. 2015. Chodatodesmus australis Sp. Nov. (Scenedesmaceae, Chlorophyta) from Antarctica, with the emended description of the genus Chodatodesmus, and circumscription of Flechtneria rotunda Gen. et Sp. Nov. Journal of Phycology 51: 1172–1188.
33.
Sherwood, A. R., Neumann, J. M., Dittbern-Wang, M. & Conklin, K. Y. 2018. Diversity of the green algal genus Spirogyra (Conjugatophyceae) in the Hawaiian Islands. Phycologia 57: 331–344.
34.
Song, G., Li, X. & Hui, R. 2017. Effect of biological soil crusts on seed germination and growth of an exotic and two native plant species in an arid ecosystem. PLoS ONE 12: 529–555.
35.
Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (* and Other Methods). Version 4. Sinauer Associates, Sunderland.
36.
Trainor, F. R., Wallett, S. & Grochowski, J. 1991. A useful algal growth medium. Journal of Phycology 27: 460–461.
37.
Trobajo, R., Mann, D. G., Clavero, E., Evans, K. M., Vanormelingen, P. & McGregor, R. C. 2010. The use of partial cox1, rbcL and LSU rDNA aequences for phylogenetics and species identification within the Nitzschia palea species complex (Bacillariophyceae). European Journal of Phycology 45: 413–425.
38.
White, J., Bruns, T. T. D., S. Lee, B. & Taylor, J. W. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322.
39.
Zhu, X., Wang, J., Lu, Y., Chen, Q. & Yang, Z. 2015. Grazer-induced morphological defense in Scenedesmus obliquus is affected by competition against Microcystis aeruginosa. Scientific Reports 5: 12743.
40.
Zou, S., Fei, C., Gao, Z., Bao, Y., He, M. & Wang, C. 2016. How DNA barcoding can be more effective in microalgae identification: a case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Scientific Reports 9: 36822.
41.
Zuccarello, G. C. & West, J. A. 2003. Multiple cryptic species: molecular diversity and reproductive isolation in the Bostrychia radicans / B. moritziana complex (Rhodomelaceae, Rhodophyta) with focus on North American isolates. Journal of Phycology 39: 948–959.
CITATIONS (14):
1.
Terrestrial Green Algae Show Higher Tolerance to Dehydration than Do Their Aquatic Sister-Species
Elizaveta Terlova, Andreas Holzinger, Louise Lewis
Microbial Ecology
Elizaveta Terlova, Andreas Holzinger, Louise Lewis
Microbial Ecology
2.
Morphology and molecular phylogeny of representatives of the genus Coelastrella Chodat from the Urals and Khentei mountain systems
Irina Novakovskaya, Irina Egorova, Nina Kulakova, Elena Patova
Issues of modern algology (Вопросы современной альгологии)
Irina Novakovskaya, Irina Egorova, Nina Kulakova, Elena Patova
Issues of modern algology (Вопросы современной альгологии)
3.
A Polyphasic Characterisation of Tetradesmus almeriensis sp. nov. (Chlorophyta: Scenedesmaceae)
Sara Turiel, Jose Garrido-Cardenas, Cintia Gómez-Serrano, Francisco Acién, Lorenzo Carretero-Paulet, Saúl Blanco
Processes
Sara Turiel, Jose Garrido-Cardenas, Cintia Gómez-Serrano, Francisco Acién, Lorenzo Carretero-Paulet, Saúl Blanco
Processes
4.
Modeling and dynamic design of an artificial culture medium for heterotrophic cultivation of Tetradesmus obliquus RDS01 for CO2 sequestration and green biofuels production: an eco-technological approach
Silambarasan Selvan, Sanjivkumar Muthusamy, Ravikumar Chandrasekaran, Dhandapani Ramamurthy, Sendilkumar Balasundaram
Biomass Conversion and Biorefinery
Silambarasan Selvan, Sanjivkumar Muthusamy, Ravikumar Chandrasekaran, Dhandapani Ramamurthy, Sendilkumar Balasundaram
Biomass Conversion and Biorefinery
5.
Effects of Dissolved Organic Matter Fractions of Varying Molecular Weights and Cd2 + on Scenedesmus obliquus Growth
V. Tikhonov, E. Voronova, M. Karpukhin, R. Aimaletdinov, V. Demin, O. Drozdova
Eurasian Soil Science
V. Tikhonov, E. Voronova, M. Karpukhin, R. Aimaletdinov, V. Demin, O. Drozdova
Eurasian Soil Science
6.
Eco-friendly approach for tannery effluent treatment and CO2 sequestration using unicellular green oleaginous microalga Tetradesmus obliquus TS03
Silambarasan Selvan, Ravikumar Chandrasekaran, Sanjivkumar Muthusamy, Dhandapani Ramamurthy
Environmental Science and Pollution Research
Silambarasan Selvan, Ravikumar Chandrasekaran, Sanjivkumar Muthusamy, Dhandapani Ramamurthy
Environmental Science and Pollution Research
7.
Green land: Multiple perspectives on green algal evolution and the earliest land plants
Richard McCourt, Louise Lewis, Paul Strother, Charles Delwiche, Norman Wickett, Vries de, John Bowman
American Journal of Botany
Richard McCourt, Louise Lewis, Paul Strother, Charles Delwiche, Norman Wickett, Vries de, John Bowman
American Journal of Botany
8.
Identification of Loliolide with Anti-Aging Properties from Scenedesmus deserticola JD052
Dae-Hyun Cho, Jin-Ho Yun, Jina Heo, In-Kyoung Lee, Yong-Jae Lee, Seunghee Bae, Bong-Sik Yun, Hee-Sik Kim
Journal of Microbiology and Biotechnology
Dae-Hyun Cho, Jin-Ho Yun, Jina Heo, In-Kyoung Lee, Yong-Jae Lee, Seunghee Bae, Bong-Sik Yun, Hee-Sik Kim
Journal of Microbiology and Biotechnology
9.
Taxonomic reinvestigation of the genus Tetradesmus (Scenedesmaceae; Sphaeropleales) based on morphological characteristics and chloroplast genomes
Hyeon Shik Cho, JunMo Lee
Frontiers in Plant Science
Hyeon Shik Cho, JunMo Lee
Frontiers in Plant Science
11.
First Insights into the Diversity of Green Algae from White Sands National Park and Organ Mountains–Desert Peaks National Monument
Karolina Fučíková, Victoria Williamson, Cameron Choquette, David Bustos, Nicole Pietrasiak
Western North American Naturalist
Karolina Fučíková, Victoria Williamson, Cameron Choquette, David Bustos, Nicole Pietrasiak
Western North American Naturalist
12.
Diversity of the scenedesmacean genus Chodatodesmus (Chlorophyta) and new cryptic species in the algae flora of Lake Baikal revealed by an integrative approach
Irina Nikolayevna Egorova, Nina Viktorovna Kulakova, Yekaterina Dzhambulatovna Bedoshvili
Organisms Diversity & Evolution
Irina Nikolayevna Egorova, Nina Viktorovna Kulakova, Yekaterina Dzhambulatovna Bedoshvili
Organisms Diversity & Evolution
13.
Diversity of algae and cyanobacteria from biological soil crusts in the high Arctic (Svalbard) along two different moisture gradients
Tatiana Mikhailyuk, Karin Glaser, Eduard Demchenko, Vivien Hotter, Ekaterina Pushkareva, Ulf Karsten
European Journal of Phycology
Tatiana Mikhailyuk, Karin Glaser, Eduard Demchenko, Vivien Hotter, Ekaterina Pushkareva, Ulf Karsten
European Journal of Phycology
14.
Versatility of Tetradesmus sp. metabolism for multi-objective culture optimization: Assessment of metabolic state in response to combined nutritional and salt stress
Nadia Berrejeb, Cecilia Faraloni, Souhir Jazzar, Giuseppe Torzillo, Issam Smaali
Algal Research
Nadia Berrejeb, Cecilia Faraloni, Souhir Jazzar, Giuseppe Torzillo, Issam Smaali
Algal Research
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.