Morphology and molecular phylogeny of Gomphonemopsis
sieminskae sp. nov. isolated from brackish waters of the
East China Sea coast
1
Palaeoceanology Unit, Faculty of Geosciences, University of Szczecin,
Mickiewicza 16a, 70–383 Szczecin, Poland
2
Key Laboratory of Marine Biotechnology of Zhejiang Province,
Ningbo University, 818 Fenghua Road, Ningbo 315211, China
3
Faculty of Biology, Institute of Ecology, Phylogeny, Diversity, J. W.
Goethe-University and Forschungungsinstitut Senckenberg, Senckenberganlage
31-33, 60054, Frankfurt am Main, Germany
4
Institute of Ecological Sciences, South China Normal University,
Guangzhou, China
Publication date: 2019-07-18
Plant and Fungal Systematics 2019; 64(1): 17-24
KEYWORDS
ABSTRACT
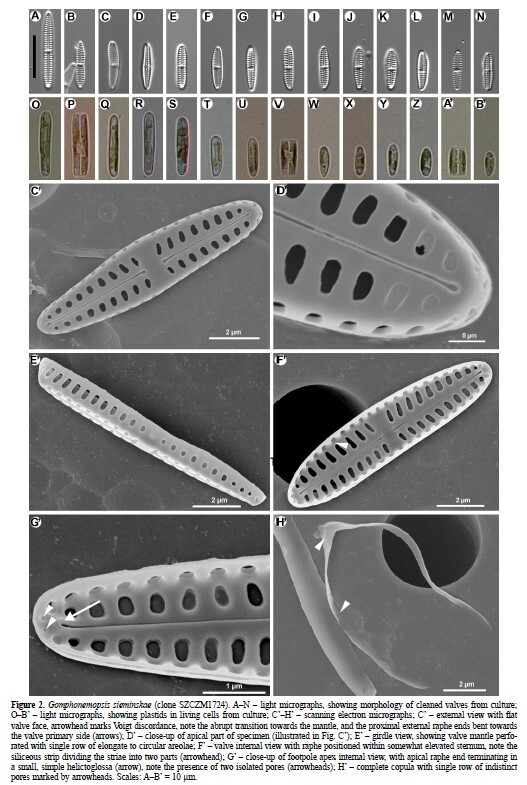
We describe the new species Gomphonemopsis sieminskae from brackish waters
of the East China Sea littoral near Ningbo, China. Two diatom strains isolated from Ulva
sp. were successfully grown, then analyzed by light (LM) and scanning electron (SEM)
microscopy. The new species is compared to known Gomphonemopsis species, and similarities
to G. pseudoexigua and G. obscurum are emphasized. Although the size metric data
overlap and the external views are fairly similar, the two taxa differ in their valve interior.
Molecular barcoding strongly discriminated G. sieminskae from G. cf. exigua, while rbcL
gene-based phylogeny showed G. sieminskae to be sister to Phaeodactylum tricornutum.
In this respect our results confirm the recent discovery, based on molecular data, that Gomphonemopsis
is a close-relative taxon to Phaeodactylum, and place it in Phaeodactylaceae
instead of Rhoicospheniaceae as inferred solely from morphology.
REFERENCES (31)
1.
Al-Handal, A., Thomas, E. W., Torstensson, A., Jahn, R. & Wulff, A. 2018. Gomphonemopsis ligowskii, a new diatom (Bacillariophyceae) from the marine Antarctic and a comparison to other Gomphonemopsis. Diatom Research 33: 97–103.
2.
Andersen, R. A, & Kawachi, M. 2005. Traditional microalgae isolation techniques. In: Andersen R. A. (ed.), Algal culturing techniques. Elsevier Academic Press: 83–100.
3.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin V. M., Nikolenko, S. I, Pham, S., Prjibelski, A. D., Pyshkin, A. V, Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A. & Pevzner, P. A. 2012. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology 19: 455–477. .
4.
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J. & Jabbari, K. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456(7219): 239–244.
5.
Chen, J. & Zhu, H. 1983. Amphiraphisales, a new order of the pennatae, Bacillariophyta. Acta Phytotaxonomica Sinica 21: 449–457.
6.
Doyle, J. J., Doyle, J. L. 1990. Isolation of Plant DNA from Fresh Tissue. Focus 12: 13–15.
7.
Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates. In: Smith, W. L. & Chanley, M. H. (eds.) Culture of marine invertebrate animals. Plenum Press: 29–60.
8.
Guslyakov, N. E. 1981. Novyi vid roda Gomphonema (Bacillariophyta) iz Chernogo Moria. Botanicheskii Zhurnal 66: 1046–1047.
9.
Guslakov, N. A., Zakordonets, I. A. & Gerasimuk, V. P. 1992. Atlas diatomovykh vodorosley bentosa severozapadnoy tschasti Tschrnogo Morja. Naukovaya Dumka.
10.
Hendey, N. I. 1977. The species diversity index of some in-shore diatom communities and its use in assessing the degree of pollution insult on parts of the north coast of Cornwall. Beihefte zur Nova Hedwigia 54: 355–378.
11.
Hinz, F., Simonsen, R., Crawford, R. M. 2012. Validation of 42 names of diatom taxa from the Baltic Sea. Diatom Research 27: 81–89.
12.
Krasske, G. 1939. Zur Kieselalgenflora Südchiles. Archiv für Hydrobiologie 35: 349–468.
13.
Kociolek, J. P., Balasubramanian, K., Blanco, S., Coste, M., Ector, L., Liu, Y., Kulikovskiy, M., Lundholm, N., Ludwig, T., Potapova, M., Rimet, F., Sabbe, K., Sala, S., Sar, E., Taylor, J., Van de Vijver, B., Wetzel, C. E., Williams, D. M., Witkowski, A. & Witkowski, J. 2019. DiatomBase. Gomphonemopsis Medlin, L. in Medlin, L. & Round, F. E. 1986. Accessed through: World Register of Marine Species at: http://www.marinespecies.org/a... on 2019-03-17.
14.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35: 1547–1549.
15.
Lange-Bertalot H., Külbs K., Lauser, T., Nörpel-Schempp M., Willmann M. 1996. Diatom taxa introduced by Georg Krasske documentation and Revision. Iconographia Diatomologica 3: 1–358.
16.
McGinnis, S., & Madden, T. L. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research 32(Web Server): W20–W25.
17.
Medlin, L. K. 1985. A reappraisal of the diatom genus Rhoiconeis and the description of Campylopyxis, gen. nov. British Phycological Journal 20: 313–328.
18.
Medlin, L. K. Round, F. E. 1986. Taxonomic studies of marine gomphonemoid diatoms. Diatom Research 1: 205–225.
19.
Metzeltin, D. & Witkowski, A. 1996. Diatomeen der Bären-Insel. Süsswasser- und marine Arten. In: Lange-Bertalot, H. (ed.), Iconographia Diatomologica. Annotated Diatom Micrographs. Vol. 4. Taxonomy. Koeltz Scientific Books: 3–217.
20.
Rombel, I. T., Sykes, K. F., Rayner, S., & Johnston, S. A. 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene 282: 33–41.
21.
Round, F. E., Crawford, R. M., Mann D. G. 1990. The diatoms: biology and morphology of the genera. Cambridge University Press.
22.
Sabir J. S. M., Theriot, E. C., Manning, S. R., Al-Malki, A., Khiyami, M. A., Al-Ghamdi, A. K., Sabir, M. J., Romanovicz, D. K., Hajrah, N. H., El Omri, A., Jansen, R. K. & Ashworth, M. P. 2018. Phylogenetic analysis and a review of history of the accidental plankter Phaeodactylum tricornutum Bohlin (Bacillariophyta). PLoS ONE 13: e0196744.
23.
Sievers, F., Wilm, A., Dineen D. G, Gibson, T. J., Karplus K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7: 539.
24.
Silva, P. C. 1962. Classification of algae. In: Lewin, R. A. (ed.), Physiology and Biochemistry of Algae, pp. 827–837. Academic Press, New York.
25.
Simonsen, R. 1959. Neue Diatomeen aus der Ostsee I. Kieler Meeresforschungen 15: 74–83.
26.
Snoeijs P. J. M. & Balashova N. 1998. Intercalibration and distribution of diatom species in the Baltic Sea. Uppsala: Opulus press 5.
27.
Snoeijs, P. & Vilbaste, S. 1994. Intercalibration and distribution of diatom species in the Baltic Sea. Uppsala: Opulus press 2.
28.
Vartanian, M., Desclés, J., Quinet, M., Douady, S., Lopez, P. J. 2009. Plasticity and robustness of pattern formation in the model diatom Phaeodactylum tricornutum. New Phytologist 182: 429–42.
29.
Witkowski, A., Lange-Bertalot, H. & Metzeltin, D. 2000. Diatom Flora of Marine Coasts I. In: Lange-Bertalot, H. (ed.), Iconographia Diatomologica. Annotated Diatom Micrographs. Diversity-Taxonomy-Identification. Koeltz Scientific Books 7.
30.
Witoń, E. & Witkowski, A. 2006. Holocene diatoms (Bacillariophyceae) from Faeroe Islands Fjords, Northern Atlantic Ocean. II. Distribution and taxonomy of marine taxa with special reference to benthic forms. Diatom Research 21: 175–215.
31.
Zhang, L., Xie, L. 2014. Modeling and Computation in Engineering III. CRC Press: 149–152.
CITATIONS (3):
1.
Chinia gen. nov.—the second diatom genus simonsenioid raphe from mangroves in Fujian, China
Huina Lin, Weiwei Wu, Lin Sun, Andrzej Witkowski, Xiaoye Li, Vishal Patil, Junrong Liang, Xuesong Li, Yahui Gao, Changping Chen
Journal of Oceanology and Limnology
Huina Lin, Weiwei Wu, Lin Sun, Andrzej Witkowski, Xiaoye Li, Vishal Patil, Junrong Liang, Xuesong Li, Yahui Gao, Changping Chen
Journal of Oceanology and Limnology
2.
Two new diatom species of the genus Gomphonemopsis (Bacillariophyceae) from the coast of China and two new combinations for the genus
Lang Li, Qun-Zhuan Nong, Chang-Ping Chen, Yu-Hang Li, Jun-Xiang Lai
PhytoKeys
Lang Li, Qun-Zhuan Nong, Chang-Ping Chen, Yu-Hang Li, Jun-Xiang Lai
PhytoKeys
3.
Benthic Marine Diatom Flora (Bacillariophyta) of Yap, Micronesia: Preliminary Annotated List, with Some New Mangrove Species
Christopher S. Lobban, Bernadette G. Tharngan
Diversity
Christopher S. Lobban, Bernadette G. Tharngan
Diversity
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.