Galbinothrix, a new monotypic genus of Chrysotrichaceae
(Arthoniomycetes) lacking pulvinic acid derivatives
1
Department of Natural History, NTNU University Museum, Erling
Skakkes gate 47a, 7012 Trondheim, Norway
2
Department of Ecology, Swedish University of Agricultural Sciences,
P.O. Box 7044, SE-750 07 Uppsala, Sweden
3
National Institute of Biological Resources, Incheon 22689, South
Korea
4
Department of Botany, National Museum of Nature and Science, 4-1-1
Amakubo, Tsukuba, 305-0005, Japan
Publication date: 2018-12-31
Plant and Fungal Systematics 2018; 63(2): 31-37
KEYWORDS
ABSTRACT
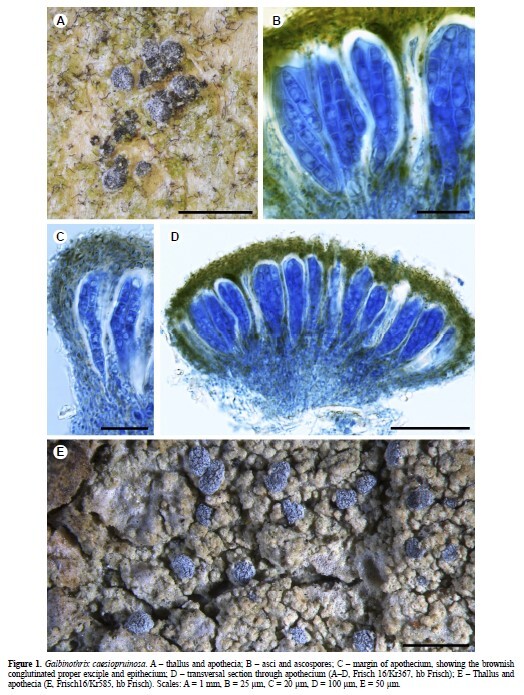
Galbinothrix caesiopruinosa is described from Japan and Korea. The new genus
and species is placed in Chrysotrichaceae by its ascoma morphology and by a phylogenetic
analysis of mtSSU and nLSU sequence data using Bayesian and maximum likelihood inference.
The monotypic genus Galbinothrix is superficially similar to Chrysothrix caesia in
having dark brown ascomata covered by a thin bluish grey pruina, reddish brown ascomatal
pigment in the epithecium and proper exciple, the greyish green to yellowish olive thallus,
and usnic acid as the main secondary thallus compound. It differs from this species and all
other Chrysotrichaceae by its large, oblong, thick-walled ascospores with a distinct epispore,
the narrowly clavate to almost tubular asci, and the never clearly granular to leprose thallus.
REFERENCES (26)
1.
Akaike, H. 1973. Information theory and an extension of the maximum likelihood principle. In: Petrov, B. N. & Casaki, F. (eds.), Second international symposium on information theory, pp. 267–281. Academiai Kiado, Budapest.
2.
Arup, U., Ekman, S., Lindblom, L. & Mattsson, J.-E. 1993. High performance thin layer chromatography (HPTLC), an improved technique for screening lichen substances. Lichenologist 25: 61–71.
3.
Elix, J. A. 2009. Chrysothricaceae. In: McCarthy, P. M. (ed.): Flora of Australia, vol. 57, Lichens 5, pp. 13–18. ABRS and CSIRO Publishing, Canberra and Melbourne.
4.
Elix, J. A. & Kantvilas, G. 2007. The genus Chrysothrix in Australia. Lichenologist 39: 361–369.
5.
Ertz, D. & Tehler, A. 2011. The phylogeny of Arthoniales (Pezizomycotina) inferred from nucLSU and RPB2 sequences. Fungal Diversity 49: 47–71.
6.
Fletcher, A. & Purvis, O. W. 2009. Chrysothrix. In: Smith, C. W., Aptroot, A., Coppins, B. J., Fletcher, A., Gilbert, O. L., James P. W. & Wolseley P. A. (eds.), The Lichens of Great Britain and Ireland, pp. 307–309. British Lichen Society, London.
7.
Frisch, A., Thor, G., Ertz, D. & Grube, M. 2014. The Arthonialean challenge: Restructuring Arthoniaceae. Taxon 63: 727–744.
8.
Grube, M. 1998. Classification and Phylogeny in the Arthoniales (Lichenized Ascomycetes). The Bryologist 101: 377–391.
9.
Holder, M. & Lewis, P. O. 2003. Phylogeny estimation: Traditional and Bayesian approaches. Nature Reviews Genetics 4: 275–284.
10.
Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310–2314.
11.
Jagadeesh Ram, T. A. M., Sinha, G. P., Lücking, R. & Lumbsch, H. T. 2006. A new species of Chrysothrix (Arthoniales: Arthoniaceae) from India. Lichenologist 38: 127–129.
12.
Kalb, K. 2001. New or otherwise interesting lichens. I. Bibliotheca Lichenologica 78: 141–167.
14.
Liu, Y. J., Whelen, S. & Hall, B. D. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808.
15.
Miadlikowska, J. & Lutzoni, F. 2000. Phylogenetic revision of the genus Peltigera (lichen-forming ascomycetes) based on morphological, chemical and large subunit nuclear ribosomal DNA data. International Journal of Plant Sciences 161: 925–958.
16.
Miller, M. A., Pfeiffer, W. & Schwartz, T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1–8. New Orleans, LA.
17.
Montagne, J. F. C. 1852. Diagnoses phycologicae. Annales des Sciences Naturelles, ser. 3, 18: 302–319.
18.
Nelsen, M. P., Lücking, R., Grube, M., Mbatchou, J. S., Muggia, L., Rivas Plata, E. & Lumbsch, H. T. 2009. Unravelling the phylogenetic relationships of lichenised fungi in Dothideomyceta. Studies in Mycology 64: 135–144.
19.
Orange, A., James, P. W. & White, F. J. 2010. Microchemical methods for the identification of lichens. 2nd edition. British Lichen Society, London.
20.
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R. P., Moret, B. M. E. & Stamatakis, A. 2009. How many bootstrap replicates are necessary? In: S. Batzoglou (ed.), Research in Computational Molecular Biology. RECOMB 2009, pp. 184–200. Lecture Notes in Computer Science 5541. Springer, Berlin, Heidelberg.
21.
Penn, O., Privman, E., Landan, G., Graur, D. & Pupko, T. 2010. An alignment confidence score capturing robustness to guide-tree uncertainty. Molecular Biology and Evolution 27: 1759–1767.
22.
Ronquist, F. & Huelsenbeck, J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinforma 19: 1572–1574.
23.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739.
24.
Vilgalys, R. & Hester, M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246.
25.
Zahlbruckner, A. 1905. Lichenes. In: H. G. A. Engler & K. A. E. Prantl (eds), Die natürlichen Pflanzenfamilien 1 (1*), pp. 97–144. Wilhelm Engelmann, Leipzig.
26.
Zoller, S., Scheidegger, C. & Sperisen, C. 1999. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31: 511–516.
CITATIONS (1):
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.